As planned, exploiting our extensive collection of cell models, we studied cancer vulnerabilities weakening the efficiency of DDR pathways. To this end, we developed a loss of function CRISPR CAS9 murine library encompassing the mammalian complement of 500 genes involved in almost all DDR pathways. To study how the loss of function of DDR genes triggers an anticancer immune response, we exploited murine CRC cell lines and syngeneic mouse models.
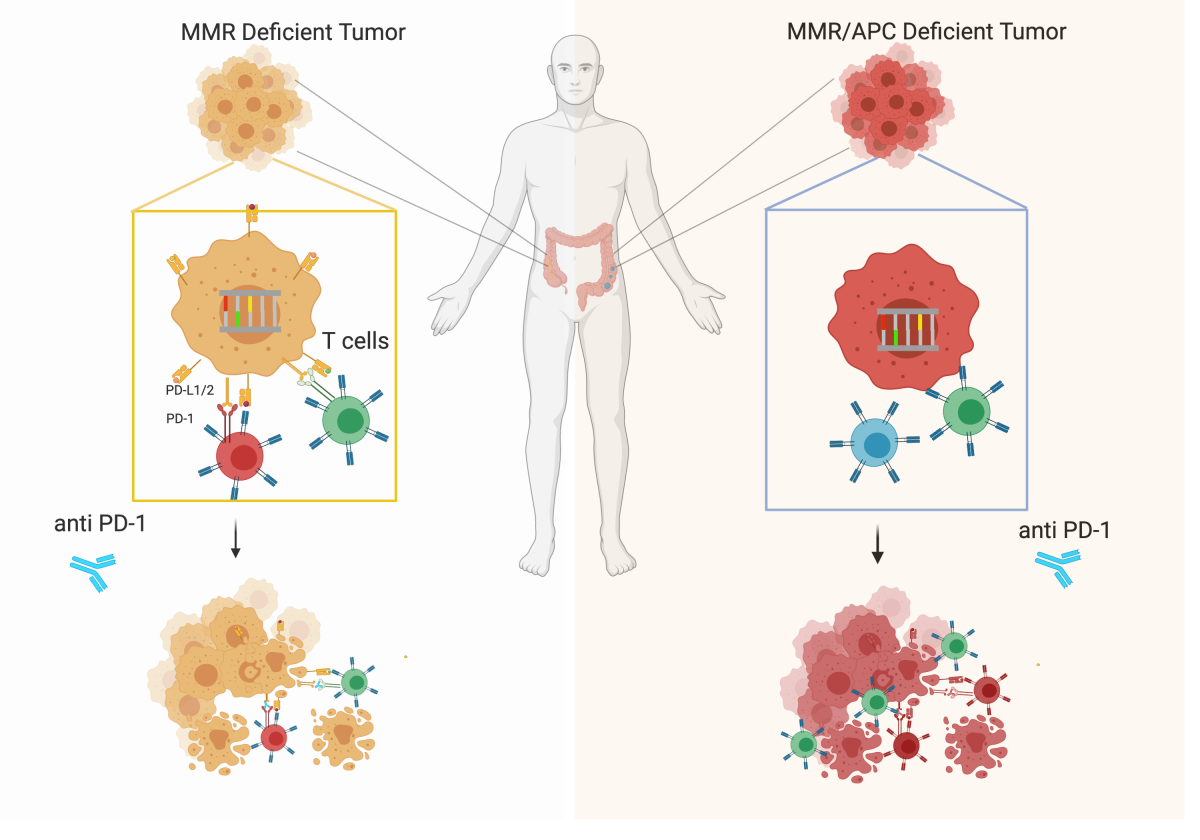
Since the majority of CRC patients are immune refractory, we assessed whether disabling DDR genes could make their tumors eligible for immune-based therapies. Recent findings show the coexistence of MSS/MMRp and MSI/MMRd cancer cells in the same tumor lesion in almost 1% of CRC patients (Fig. 3).
Intrigued by these evidences, we generated a preclinical model of MMR heterogeneity, mixing Mlh1+/+ and Mlh1-/- colorectal cancer murine cells at different ratio. Notably, tumor growth delay and tumor rejections occurred in Mlh1+/+ and Mlh1-/- mixed tumors and increased when the percentages of Mlh1-/- cells augmented in the mixed population. The observation that a MMRp tumor harboring a small fraction of MMRd cells can trigger an effective antitumor immune response has implications for the rational design of clinical trials for tumors recalcitrant to immune checkpoint blockade (ICB).
Tumors can evade immune surveillance through alterations in the antigen presenting machinery (APM). Several studies reported that molecular defects in the major complex of histocompatibility I (MHC I) and in the protein Beta 2 Microglobulin (B2M) represent mechanisms of acquired resistance to ICB in melanoma and lung tumors. We evaluated the impact of B2M loss in immune evasion and resistance to ICB in colorectal, pancreatic and breast cancer murine cell lines. Although the antigen presentation was compromised, the growth of MMRd murine cell lines was still severely impaired by the administration of ICB. The analysis of tumor microenvironment revealed that CD4+ T cells were pivotal in establishing an effective cancer immune response but only in the context of MMRd tumors (Fig. 4).
The impact of the non-coding DNA (98% of the entire genome) on the immunogenicity of MMRd tumors is largely unknown. As part of TARGET we recently found that non-canonical transcripts, can be sources of immunogenic peptides in murine CRC cells (Rospo et al., Genome Medicine, 2024). We initially identified the non-coding derived neoantigens that bound the MHC class I complex and then identified those that were lost after immune pressure in immune-competent mice (the same neoantigens were preserved in immune-compromised mice). Finally, we validated the immunogenicity of the peptides through the measurement of IFN-γ release from splenocytes co-cultured with synthetic peptides. Overall, we provided proof-of-concept that in MMRd tumors, the non-coding part of the genome can effectively contribute to defining the immunogenic properties of these tumor types.
As conceived, TARGET relies on the support of NeoPhore (https://www.neophore.com/) that aims to discover whether the DNA MMR machinery could be modulated with a small molecule inhibitor, thereby enabling studies on the time-dependent sequelae of MMR deficiency. In collaboration with NeoPhore we found that resorcinol fragment-derived inhibitor 1 (NP1867) binds irreversibly with high affinity and inactivation rate to PMS2 through adduction of Cys73 in the ATP binding site, as demonstrated by protein-ligand X-ray crystallography. In vitro and cell-based selectivity profiling of NP1867 revealed minimal off-target activity. Importantly, in cell-based mechanistic and functional assays, the inhibition of PMS2 by NP1867 was observed.
Expected results until the end of the project
We had previously reported that inactivation of DNA MMR leads to MSI status and generates hypermutated cancers with increased number of neoantigens. We also found that treatment of mouse and human CRC cells with temozolomide (TMZ) leads to MMR deficiency, increasing tumor mutational burden (TMB) and response to ICB. Altogether these preclinical data led to design ARETHUSA, a proof-of-concept two steps clinical trial (Fig. 5).
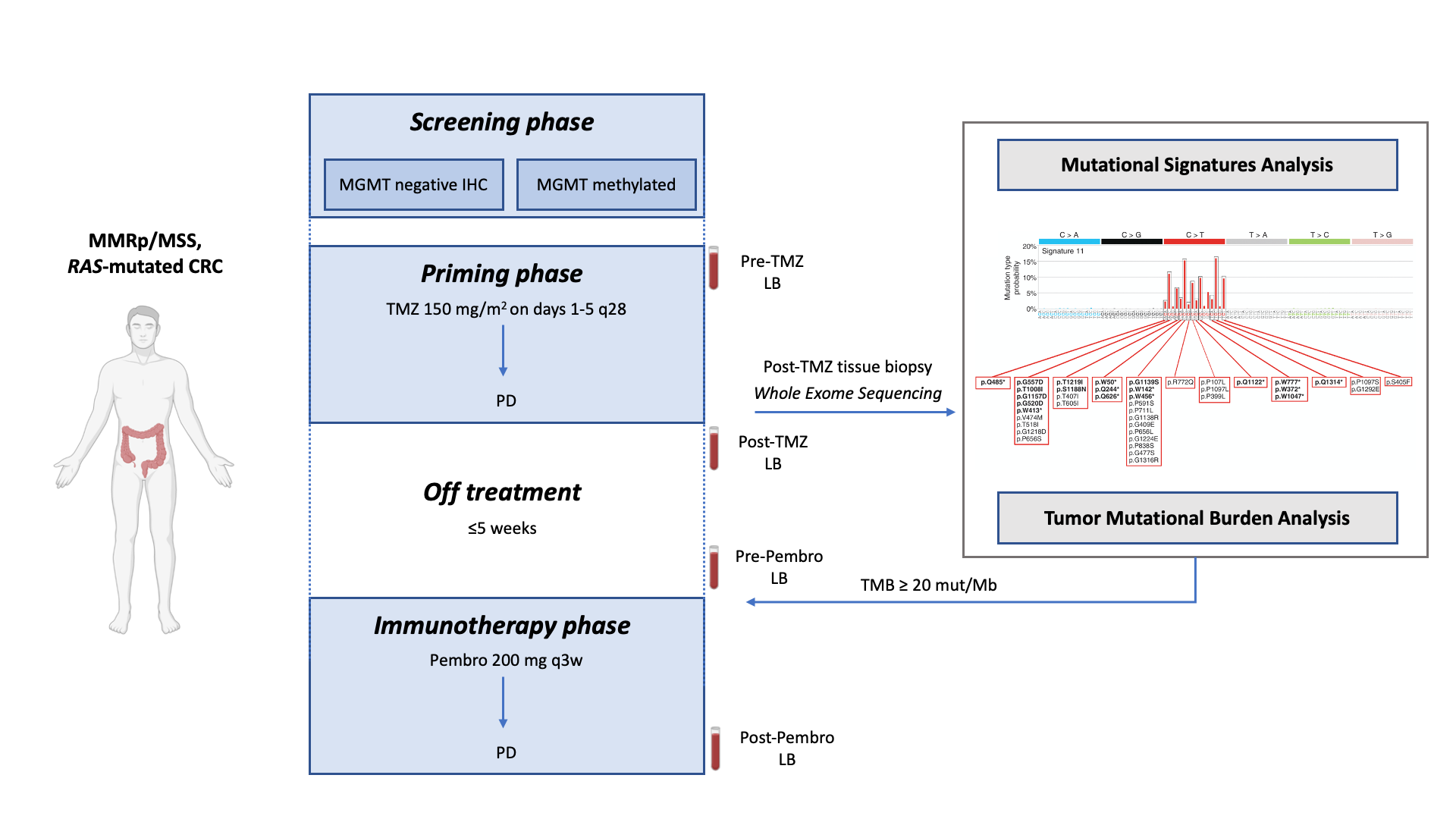
During the first step the TMZ treatment was used both with curative intent and to trigger an hypermutation status in MSS mCRC patients. In the second step the anti PD-1 agent pembrolizumab was deployed only if patients develop a TMB > 20 mut/Mb upon progression to TMZ. Furthermore, in patients receiving a prolonged treatment of TMZ, alterations in MMR emerged. Interestingly, we found that a subset of the patients whose tumors displayed the acquisition of MMR deficiency and increased TMB, achieved disease stabilization upon pembrolizumab treatment.
Overall, these results indicate that pharmacological agents can be used to disable DNA repair pathways, and this can lead to clinical benefit in mCRC patients refractory to immune based treatments.
Publications
- Mariella E, Grasso G, Miotto M, Buzo K, Reilly NM, Andrei P, Vitiello PP, Crisafulli G, Arena S, Rospo G, Corti G, Lorenzato A, Cancelliere C, Barault L, Gionfriddo G, Linnebacher M, Russo M, Di Nicolantonio F, Bardelli A.
Transcriptome-wide gene expression outlier analysis pinpoints therapeutic vulnerabilities in colorectal cancer.
Mol Oncol. 2024 Mar 11. [Online ahead of print] - Sogari A, Rovera E, Grasso G, Mariella E, Reilly NM, Lamba S, Mauri G, Durinikova E, Vitiello PP, Lorenzato A, Avolio M, Piumatti E, Bonoldi E, Aquilano MC, Arena S, Sartore-Bianchi A, Siena S, Trusolino L, Donalisio M, Russo M, Di Nicolantonio F, Lembo D, Bardelli A.
Tolerance to colibactin correlates with homologous recombination proficiency and resistance to irinotecan in colorectal cancer cells
Cell Rep Med. 2024 Feb 20;5(2):101376 - Mauri G, Patelli G, Roazzi L, Valtorta E, Amatu A, Marrapese G, Bonazzina E, Tosi F, Bencardino K, Ciarlo G, Mariella E, Marsoni S, Bardelli A, Bonoldi E, Sartore-Bianchi A, Siena S.
Clinicopathological characterisation of MTAP alterations in gastrointestinal cancers.
J Clin Pathol. 2024 Feb 13:jcp-2023-209341. [Online ahead of print] - Maione F, Oddo D, Galvagno F, Falcomatà C, Pandini M, Macagno M, Pessei V, Barault L, Gigliotti C, Mira A, Corti G, Lamba S, Riganti C, Castella B, Massaia M, Rad R, Saur D, Bardelli A, Di Nicolantonio F.
Preclinical efficacy of carfilzomib in BRAF-mutant colorectal cancer models.
Mol Oncol. 2024 Feb 13.[Online ahead of print] - Rospo G, Chilà R, Matafora V, Basso V, Lamba S, Bartolini A, Bachi A, Di Nicolantonio F, Mondino A, Germano G, Bardelli A.
Non-canonical antigens are the largest fraction of peptides presented by MHC class I in mismatch repair deficient murine colorectal cancer
Genome Med. 2024 Jan 19;16(1):15. - Di Nicolantonio F, Bardelli A
Precision oncology for KRASG12C-mutant colorectal cancer.
Nat Rev Clin Oncol. 2023 Jun;20(6):355-356 - Amodio V, Lamba S, Chilà R, Cattaneo CM, Mussolin B, Corti G, Rospo G, Berrino E, Tripodo C, Pisati F, Bartolini A, Aquilano MC, Marsoni S, Mauri G, Marchiò C, Abrignani S, Di Nicolantonio F, Germano G, Bardelli A.
Genetic and pharmacological modulation of DNA mismatch repair heterogeneous tumors promotes immune surveillance.
Cancer Cell. 2023 Jan 9;41(1):196-209. - Pergolizzi M, Bizzozero L, Maione F, Maldi E, Isella C, Macagno M, Mariella E, Bardelli A, Medico E, Marchiò C, Serini G, Di Nicolantonio F, Bussolino F, Arese M.
The neuronal protein Neuroligin 1 promotes colorectal cancer progression by modulating the APC/β-catenin pathway.
J Exp Clin Cancer Res. 2022 Sep 2;41(1):266. - Durinikova E, Reilly NM, Buzo K, Mariella E, Chilà R, Lorenzato A, Dias JML, Grasso G, Pisati F, Lamba S, Corti G, Degasperi A, Cancelliere C, Mauri G, Andrei P, Linnebacher M, Marsoni S, Siena S, Sartore-Bianchi A, Nik-Zainal S, Di Nicolantonio F, Bardelli A*, Arena S* (* Shared last authorship)
Targeting the DNA Damage Response Pathways and Replication Stress in Colorectal Cancer.
Clin Cancer Res. 2022 Sep 1;28(17):3874-3889. - Russo M, Pompei S, Sogari A, Corigliano M, Crisafulli G, Puliafito A, Lamba S, Erriquez J, Bertotti A, Gherardi M, Di Nicolantonio F, Bardelli A, Cosentino Lagomarsino M.
A modified fluctuation-test framework characterizes the population dynamics and mutation rate of colorectal cancer persister cells.
Nat Genet. 2022 Jul;54(7):976-984. - Crisafulli G, Sartore-Bianchi A, Lazzari L, Pietrantonio F, Amatu A, Macagno M, Barault L, Cassingena A, Bartolini A, Luraghi P, Mauri G, Battuello P, Personeni N, Zampino MG, Pessei V, Vitiello PP, Tosi F, Idotta L, Morano F, Valtorta E, Bonoldi E, Germano G, Di Nicolantonio F, Marsoni S, Siena S, Bardelli A.
Temozolomide treatment alters mismatch repair and boosts mutational burden in tumor and blood of colorectal cancer patients.
Cancer Discov. 2022 Jul 6;12(7):1656-1675.
